OVERVIEW
The "Nano Pill Bug" we are developing is a device designed to protect unmodified miRNAs from degradation by RNase. This device is inspired by pill bugs, and when it captures miRNA, it rolls up and protects it.
This structure consists of two DNA origami sheets and several stopper DNAs which help maintain the integrity of the structure. One sheet has a 2-layered structure, which bends due to the difference in the number of base pairs, forming a tubular structure.
These bent sheets are held together by pre-arranged stopper DNAs, maintaining a flat structure by balancing the tension between them. When the stopper DNA is replaced by miRNA through strand displacement, the tension in each sheet is released, and the miRNA is enclosed inside the tubular structure.
The functionality of this structure can be extended, and by applying it to small molecules such as DNA or proteins, various applications are expected.
PRINCIPLE
1. Bending DNA origami
4Helix bundles typically exhibit a linear shape. (Fig.1,2)
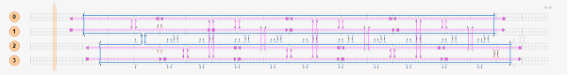
Fig.1 Modeling of linear 4Helix bundles
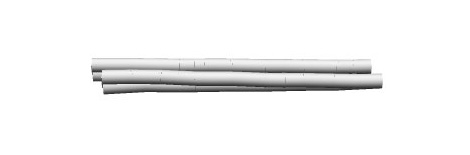
Fig.2 Simulation of the Structure
The linear structure can be changed to a bent structure by adding a base insert to one of the two upper and lower strands of the 4-helix and a base skip to the other.(Fig.3,4)
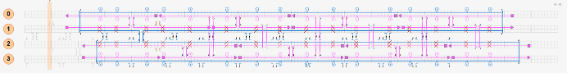
Fig.3 Modeling of bent 4Helix bundles

Fig.4 Simulation of the Structure
Also, the number of skips and inserts can be adjusted to change the curvature at will. Using this mechanism, the Nano Pill Bug transforms a flat sheet structure made of a single scaffold chain into a rounded, tubular sheet structure.
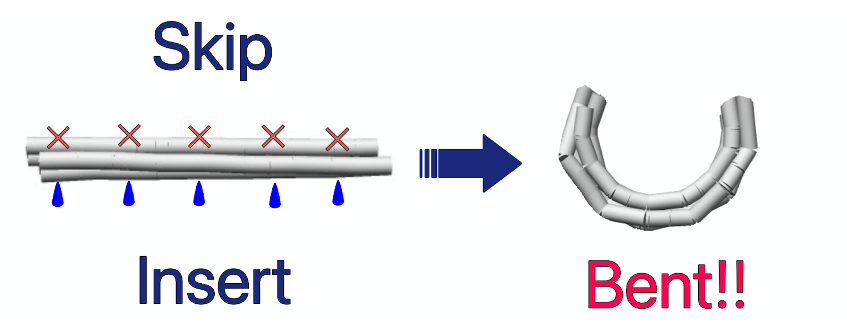
Fig.5 How DNAorigami is bent
2. Strand Displacement Reaction
A strand displacement reaction is a reaction between two DNA (or RNA) strands or between DNA and RNA in which one strand is replaced by another complementary single-stranded DNA.
By utilizing this phenomenon, the target molecule becomes a signal and the two sheets are separated into one sheet, each of which supplements the target molecule, and the sheet curls up to protect it from RNase.
Fig.6 Strand Displacement Reaction
MODELING
1. Modeling of a Single Rolled Sheet Structure
1.1. Determination of sheet size
The sheet structure was designed using caDNAno. The Nano Pill Bug consists of a single scaffold chain and 226 staple chains (average 31 nt). The scaffold chain was designed using M13mp18 (7249 nt). (Fig.7)
Fig.7 Modeling of Single Sheet Structure in caDNAno
In order for the Nano Pill Bug to more precisely protect miRNAs supplemented from RNase, the basic structure of the Nano Pill Bug, the sheet structure, is now a two-layer structure with 4-helix bundles arranged side by side. The aspect ratio of the rectangular sheet was also determined to protect fewer input miRNAs while keeping the radius small.
Finally, the sheet was determined to be 36 nm in length (18 dsDNA strands) and 68 nm in width (200 nt). (Fig.8)

Fig.8 Single Rolled Sheet Size and Structure
1.2. Determine Skip/Insert frequency
We designed the structure of the Nano Pill Bug by bending the sheets by adding a skip to one of the two designed sheets and an insert to the other, so that the ends used as the adhesive between the two sheets would not be hidden by wrapping the sheets when designing the two back-to-back structure. We also decided to “wrap the sheet a little more than once” to protect the miRNAs from the RNase.
As a result, a total of 25 skips or inserts were added to the 200 nt long sheet to “wrap the sheet a little more than once”. Of these 25 skips or inserts, 22 were set uniformly, avoiding crossover areas in the sheet.
In order to design the sheet so that only one end is bent more strongly and inward at the overlap of the sheet ends, the remaining three inserts were set to overlap the three skip inserts from one end of the sheet to the other. (Fig.9)
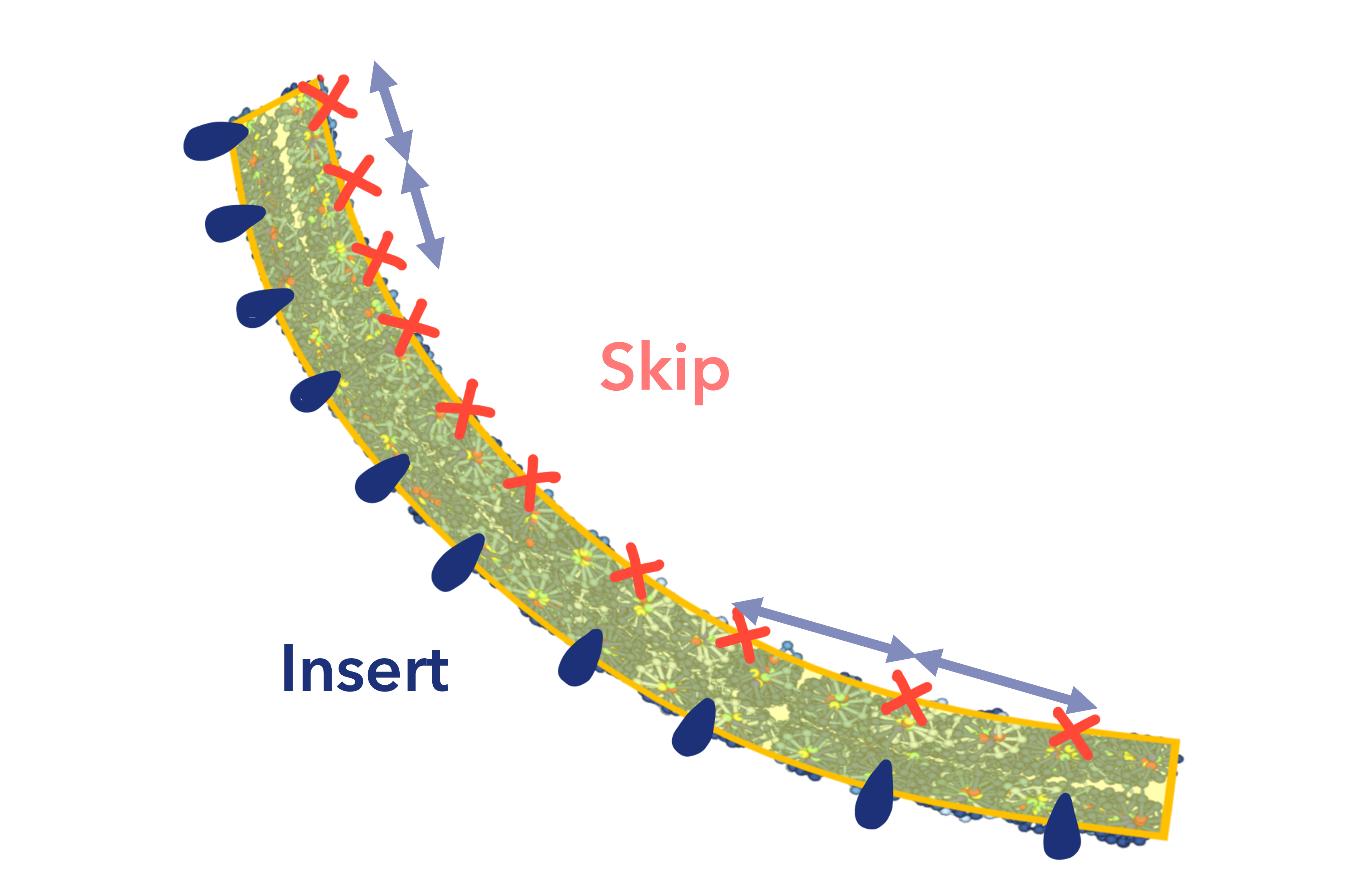
Fig.9 Relationship between Skip/Insert frequency and DNAorigami bending
2. Modeling of Back-to-back Construction of Double Sheets
Design of signal response mechanism using chain displacement reaction
NUPACK was used to design the sequence of the strand displacement reaction and to simulate the strand displacement reaction.
The method used to join the two sheets together was to use ssDNA to join the staple strands that make up the ends of the sheets together. (Fig.10)
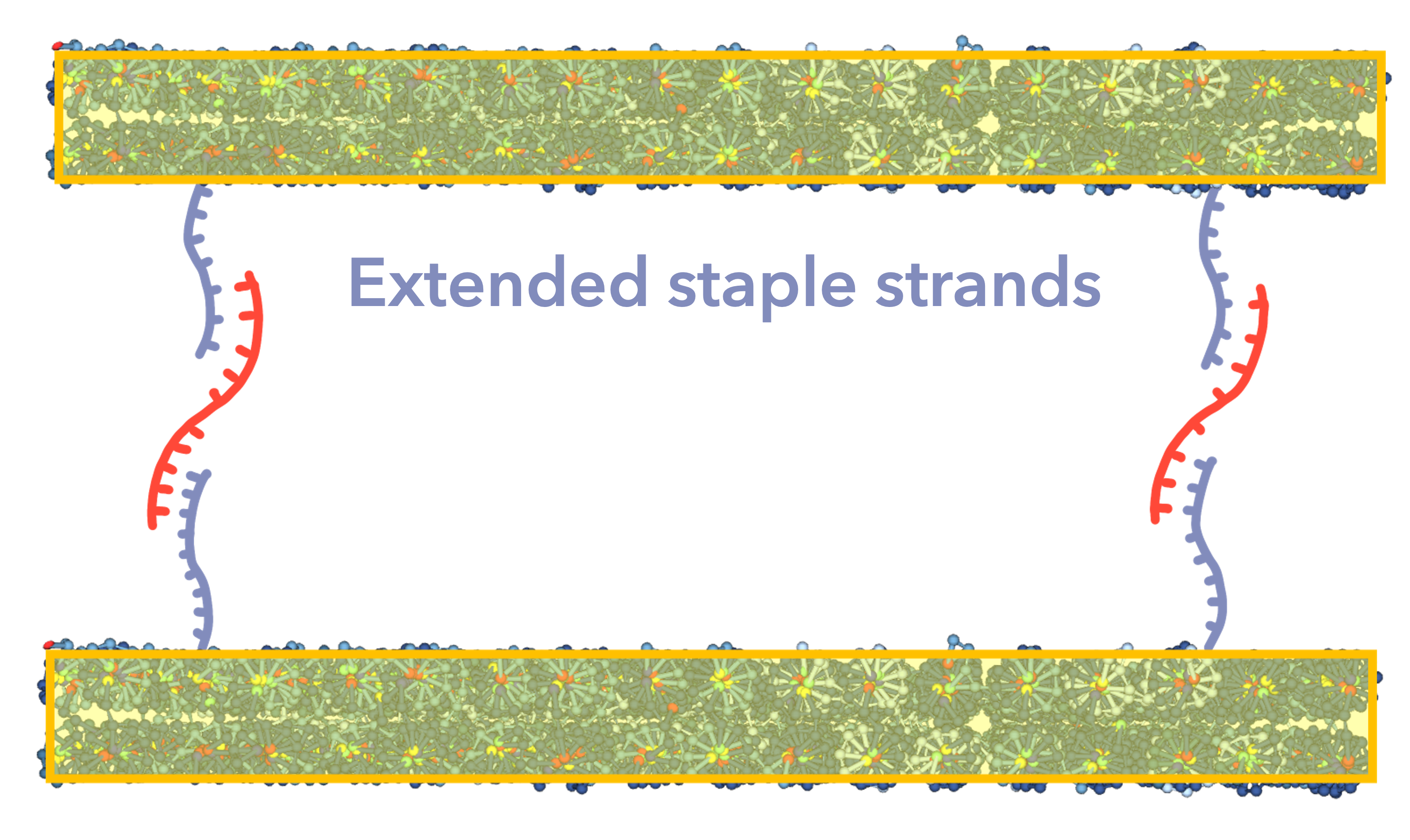
Fig.10 Connection of Double Sheets
The sheets are glued back-to-back in such a way that the bending stress of each sheet cancels each other out, resulting in a two-sheet structure fixed to a flat surface. (Fig.11)
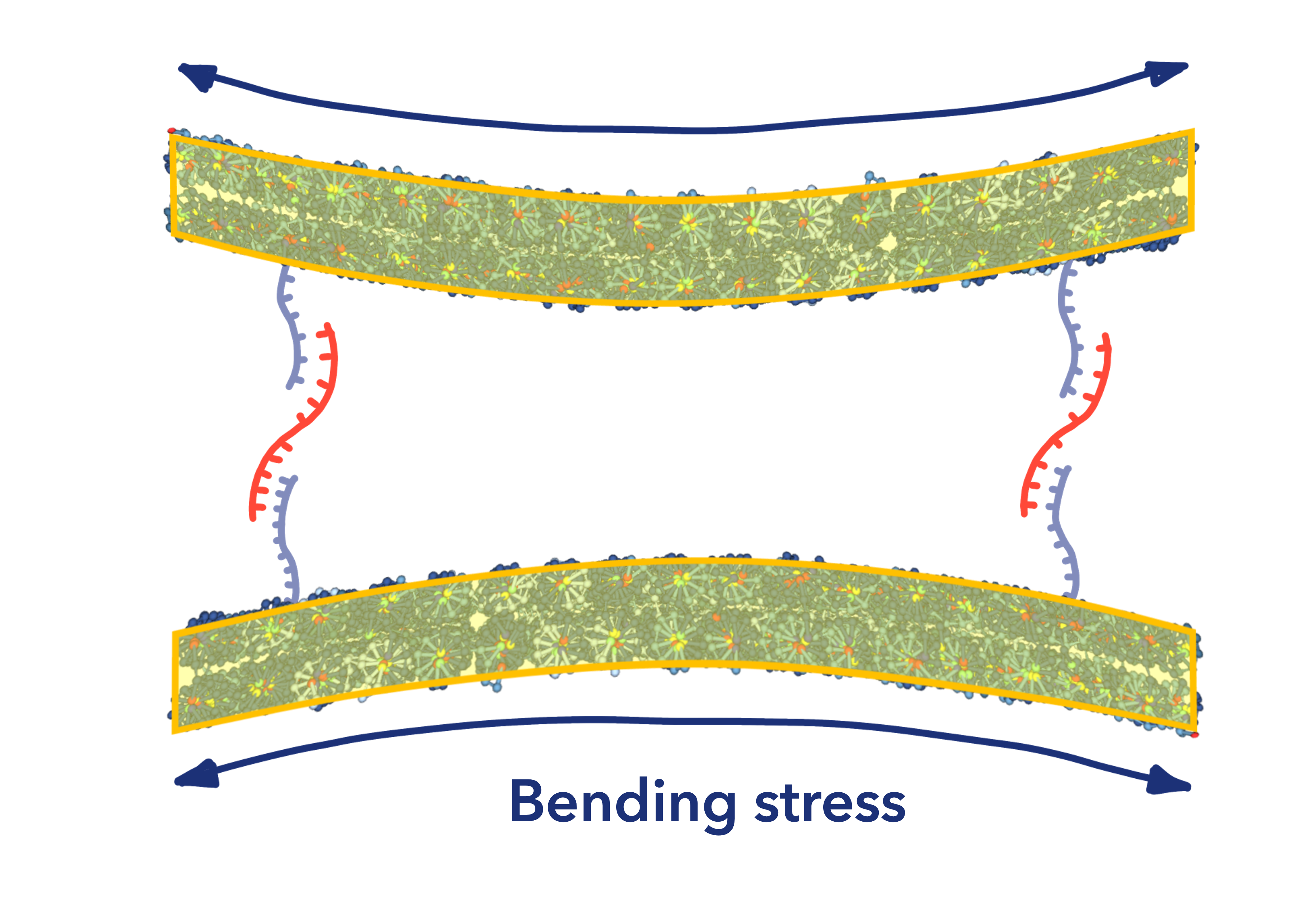
Fig.11 Back-to-back Construction of Double Sheets
When miRNAs (Target) hybridize with the end of the staple chain through strand displacement reaction, the back-to-back sheets are removed from each other, and at the same time, the sheets are rounded by the bending stress of the sheets to protect the miRNAs attached to the end of the staple chain. The sheets are then disentangled from each other. (Fig.8)
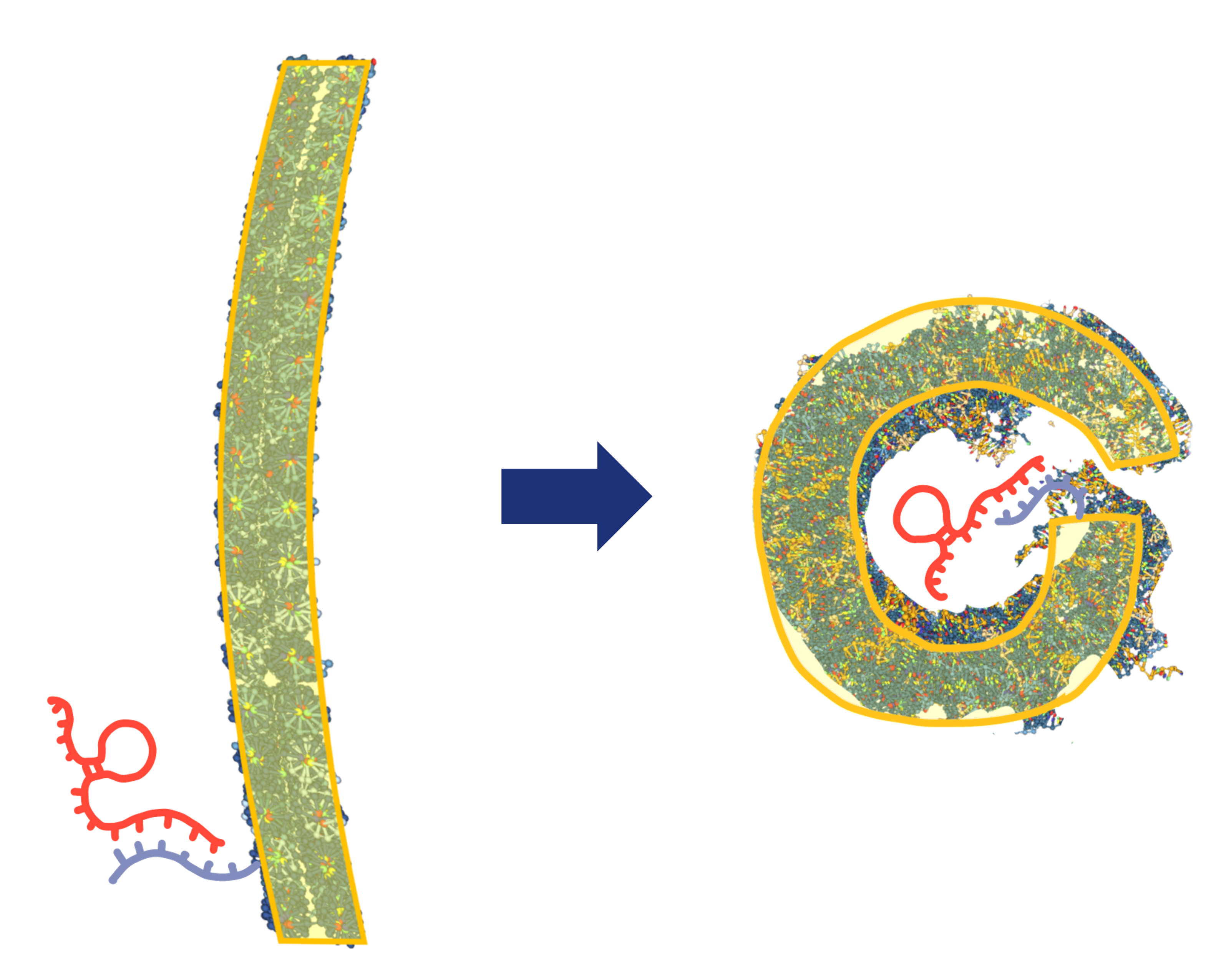
Fig.12 DNA origami that curls up in response to miRNA.
The sequence of the strand displacement reaction portion was determined using NUPACK as shown below. (Fig.13)
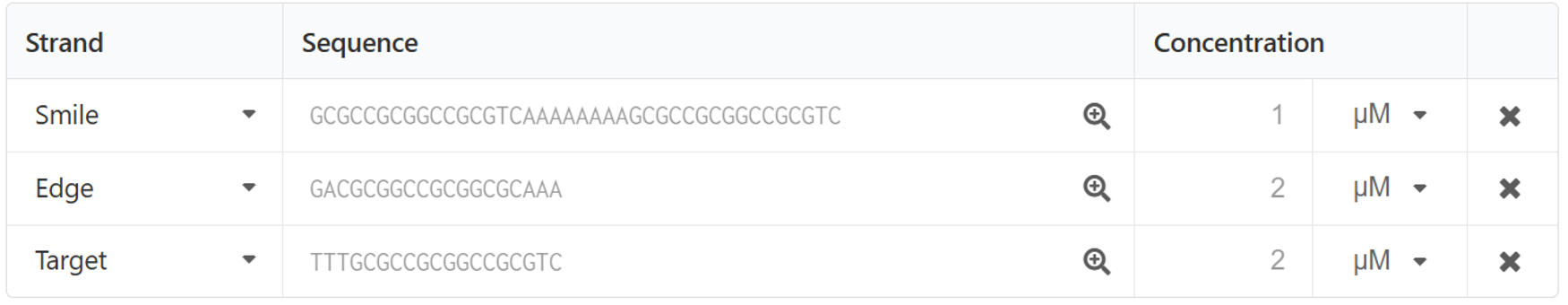
Fig.13 The sequence of this strand displacement reaction
Our Challenge
Stepwise synthesis of DNA origami
Although we have discussed the design of a two-sheet design, there is one problem in its actual synthesis.
When the two sheets are synthesized, there is one problem: if two sheets are synthesized at the same time, they are synthesized separately. Since each sheet has strong bending stress, they curl up with the connection part inside. As a result, the two sheets are synthesized in a split state, and the desired back-to-back two sheets cannot be synthesized.
To solve this problem, we considered stepwise synthesis of DNA oriami. First, the scaffold strand is joined to the connecting Staple strand, and then the remaining Staple strand is added. This allows the first connection to resist bending stress, and the desired two back-to-back sheets can be synthesized.
Improved protection from RNase
The current model has gaps on its lateral surfaces, which do not fully prevent RNase from penetrating the structure.
The first possible solution is to attach a cholesterol-modified staple chain to the surface of the sheet. This would not only electrically inhibit RNase penetration, but also preserve the bending stress of the original DNA Origami, which is an extension of our project and easy to demonstrate. Other ideas we are considering include stacking the sheets more to make them more airtight and to reduce their radius, as well as increasing the bending stress at the edges of the sheets and making the sheets more spherical.
We believe that these ideas are effective means of making nano-dangling worms a more practical technology.
- [1]Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009 Aug 7;325(5941):725-30. doi: 10.1126/science.1174251. PMID: 19661424; PMCID: PMC2737683.
- [2]Douglas SM, Marblestone AH, Teerapittayanon S, Vazquez A, Church GM, et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009
- [3]CanDohttps://cando-dna-origami.org/#:~:text=CanDo%20%E2%80%93%20Computer-aided%20engineering%20forhttps://cando-dna-origami.org/#:~:text=CanDo%20%E2%80%93%20Computer-aided%20engineering%20for